Dr. Shih-Hsun Chen

- Journal Papers
- Degrees and Positions Held
- Research
| School Name | Department | Degree | Period |
| National Taiwan University | Institute of Biochemical Sciences | Assistant Professor | 2020 - present |
| City of Hope | Postdoctoral Research Fellow | 2015 - 2020 | |
| University of Michigan, Ann Arbor | Postdoctoral Research Fellow | 2014 - 2015 | |
| Academia Sinica | Institute of Biological Chemistry | Postdoctoral Research Fellow | 2014 - 2014 |
| National Chiao Tung University | Department of Biological Science & Technology | Ph.D. | 2014-01 |
Research interests of our laboratory
Our research interests focus on exploring the mechanisms of DNA repair and treatments of DNA repair-related diseases (such as cancer, aging, neurodegenerative diseases, etc.) Based on mechanistic studies, we will identify small-molecule drugs or novel strategies for the treatments of human diseases. The topics are divided into four directions:
DNA repair is closely related to human diseases
DNA damage often occurs in the cells of every person's body. What causes DNA damage in our cells? There are two main sources, one is exogenous factors (such as radiation, viral infection, and chemicals), the other is endogenous stresses that are from replication errors and cellular metabolism (such as reactive oxygen species). Once DNA damage occurs, it will affect cell cycle, transcription, DNA repair, and cell apoptosis. Because lots of single-stranded and double-stranded DNA will break in each cell cycle, even in every minute, and DNA damage involves in some important biological processes, it is associated with many human diseases, such as immunological disorders, cancer, aging, and neurological disorders. Thus, DNA repair is very important when DNA damage occurs. DNA repair defects will cause gene instability and the occurrence of diseases (Figure 1)..jpg)
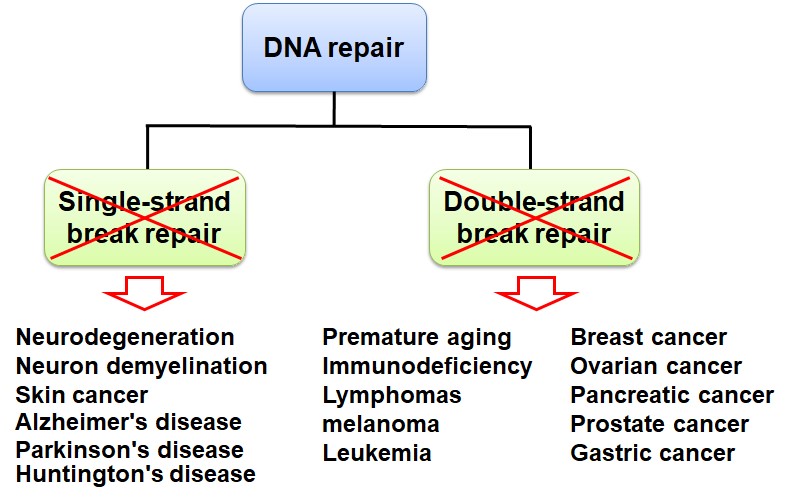
Figure 1. Loss of DNA repair causes many human diseases, including tumorigenesis, aging, neurodegenerative diseases etc.
Post-translational modifications are important for DNA repair
Post-translational modifications, such as protein phosphorylation, ADP-ribosylation, ubiquitination, acetylation, and methylation, are important for numerous biological processes, including DNA damage response. It has been reported that protein phosphorylation, ADP-ribosylation, and ubiquitination facilitate DNA single-strand break (SSB) and/or double-strand break (DSB) repair. Numerous studies investigating ADP-ribosylation have been performed in the last several decades. Yet, our understanding of the molecular mechanisms governing ADP-ribosylation signaling and the physiological and pathophysiological importance of the pathways regulated by ADP-ribosylation are still poorly understood. Thus, there are many exciting findings waiting to be discovered in this field of research. For example, the enzymatic activities of “ADP-ribosylation erasers” have been studied but their biological functions in DNA damage response are still unclear (Figure 2). Moreover, the relationship between ubiquitination, acetylation or methylation and DNA repair is also worth further investigating.
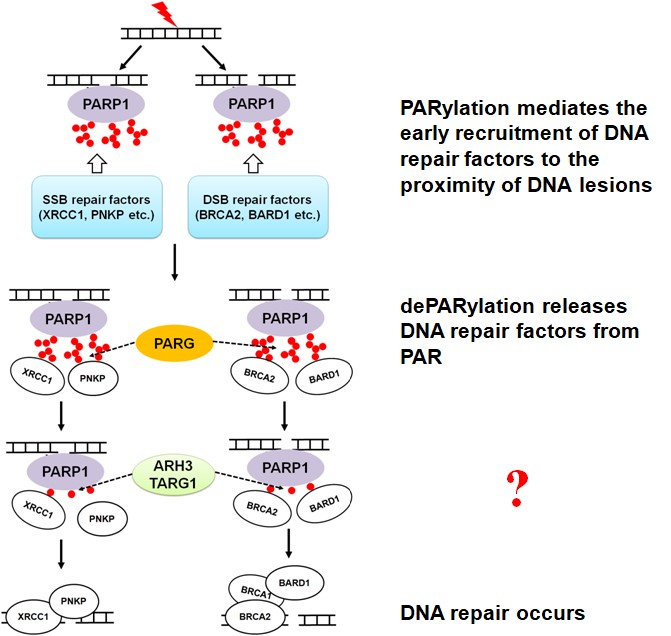
Figure 2. mono- and poly-ADP-ribosylation play an important role for DNA repair
Mechanism-based drug discovery for DNA repair-related human diseases
Our previous study shows that BRCA-deficient breast and ovarian cancer cells are hypersensitive to the treatment of PARG inhibitor, COH34 (Figure 3). Thus, similar to PARP and PARG inhibitor, small molecules that suppress DNA repair are able to be utilized for cancer chemotherapy. In particular, PARP inhibitor has been applied in the clinical treatment of cancers with BRCA mutation. Thus, we plan to examine if DNA repair-defective breast, ovarian, lung, colon, and pancreatic cancers are hypersensitive to these small molecules identified by our in silico strategies. In addition to cancer, DNA repair is also associated with aging and neurodegenerative diseases. The new drug targets and small-molecule regulators for anti-aging and neurodegenerative diseases will be identified based on mechanistic studies. We expect that our studies could be applied for clinical treatments in the future.
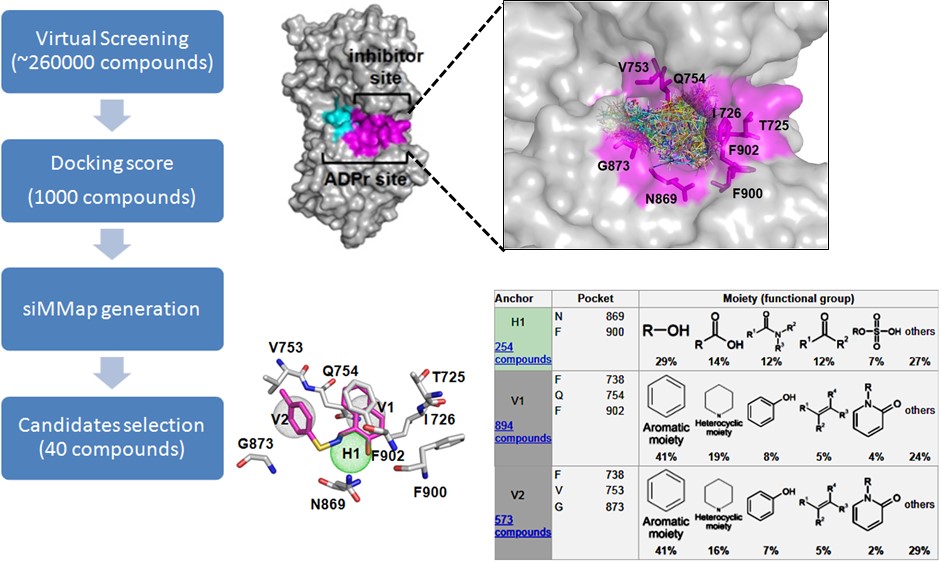

Figure 3. Using in silico strategy with siMMap analysis to identify novel PARG inhibitor (COH34) for DNA repair-defective cancer treatments.
